Ever tried searching for medical papers using a standard search engine? Happy with results you get? Probably not. There are serious limitations to using keyword search in the pharmaceutical industry.
Imagine you were looking for papers featuring the enzyme type ‘GSK’. Using a generic search engine, you would get articles that mentioned ‘glycogen synthase kinase’, as well as articles about the company ‘GlaxoSmithKline’ – which is not particularly relevant to your search here. However, a semantic search engine, powered by scientific vocabularies and a disambiguation system, will just focus on results featuring the protein, giving you context specificity.
If you needed even more accuracy and wanted to find a specific protein such as GSK3, you would be required to do a search for:
glycogen synthase kinase 3 alpha, GSK-3-A, GSK3A, alpha glycogen synthase kinase-3, glycogen synthase kinase-3A…
It’s a pretty long list of synonym derivatives, right? A good semantic search system on the other hand, does all this for you when it indexes, so that you don’t have to worry when searching.
Transformative Data Integration
Having done this, you are then set up for better downstream data analysis because your conversion from unstructured to structured (typed) data is way more accurate.
You can then connect your enriched, structured data to databases and other systems, giving enhanced data connectivity across the organisation and speeding up analysis.
Group Level Searches
Great semantic search provides taxonomic relationships between its entities, so higher order searches are possible. Let’s take the example of ‘Viagra’ – whose current use was found as an adverse effect during its trials for pulmonary hypertension.
I’d find a bunch of articles that would mention things like Viagra’s protein target, Phosphodiesterase 5A (PDE5A). The image below shows how PDE5A and Phosphodiesterase 11A (PDE11A) were found in an article and where they sit in the taxonomy.
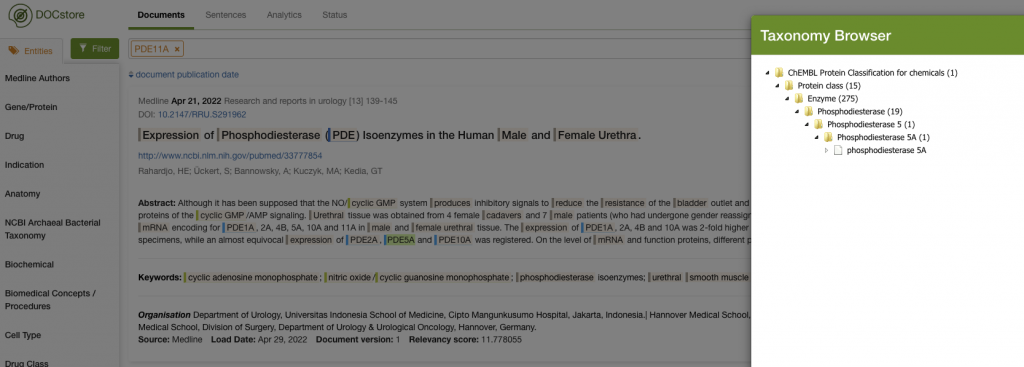 We can see that PDE5A sits in an enzyme taxonomy under the wider ‘Phosphodiesterase’ class. I could click on the ‘Phosphodiesterase’ class and get the system to search for anything under it:
We can see that PDE5A sits in an enzyme taxonomy under the wider ‘Phosphodiesterase’ class. I could click on the ‘Phosphodiesterase’ class and get the system to search for anything under it:
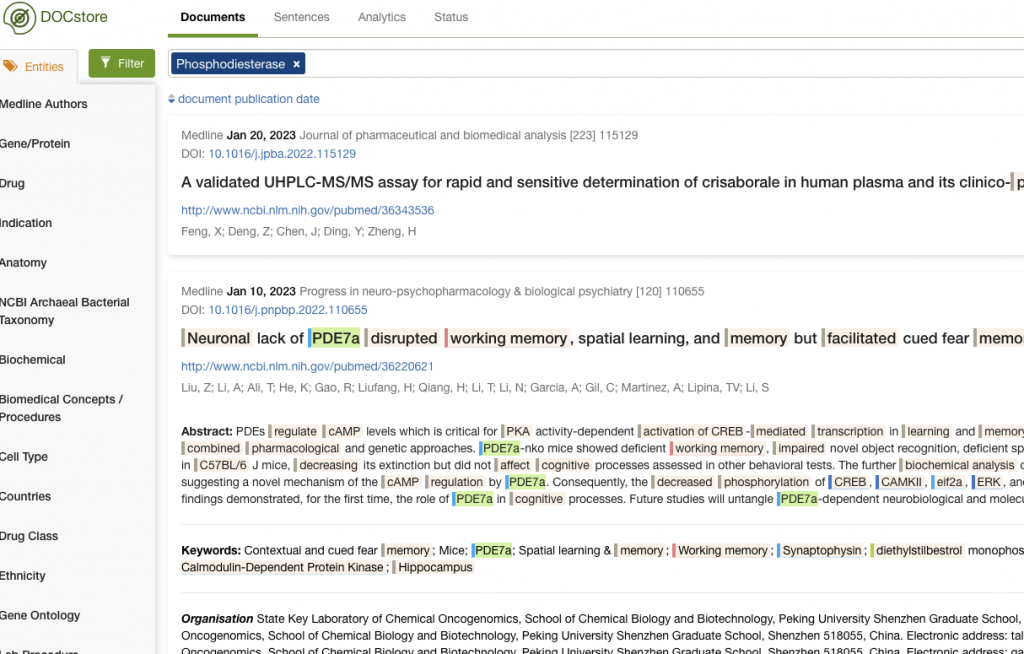 You can see how PDE8B and PDE10A were identified in this way.
You can see how PDE8B and PDE10A were identified in this way.
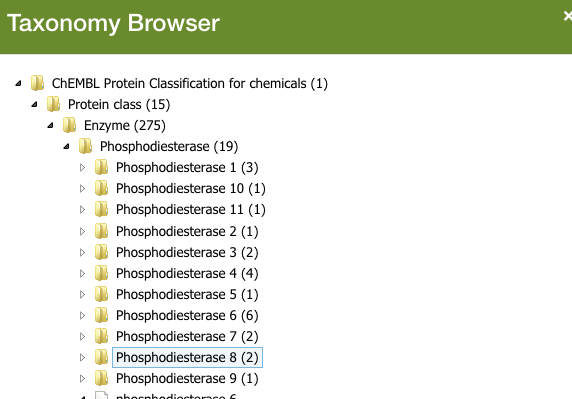 This becomes incredibly useful, say if you’re interested in finding out which competitors have developed drugs for a target you’re working on.
This becomes incredibly useful, say if you’re interested in finding out which competitors have developed drugs for a target you’re working on.
What you’re looking for is a rich set of taxonomies covering areas such as diseases, drugs, protein classes and so on.
A good semantic search engine will actually embed the concepts (that’s to say, entities such as “PDE5A”, entity classes, e.g. “gene”, or higher level abstractions like “protein class”) within the plain text. How is this useful? Well, query time is really quick and extremely accurate, all because you don’t have to do synonym expansion.
That’s hard to do in a generic search engine that doesn’t leverage life science taxonomy data in one step.
Connections
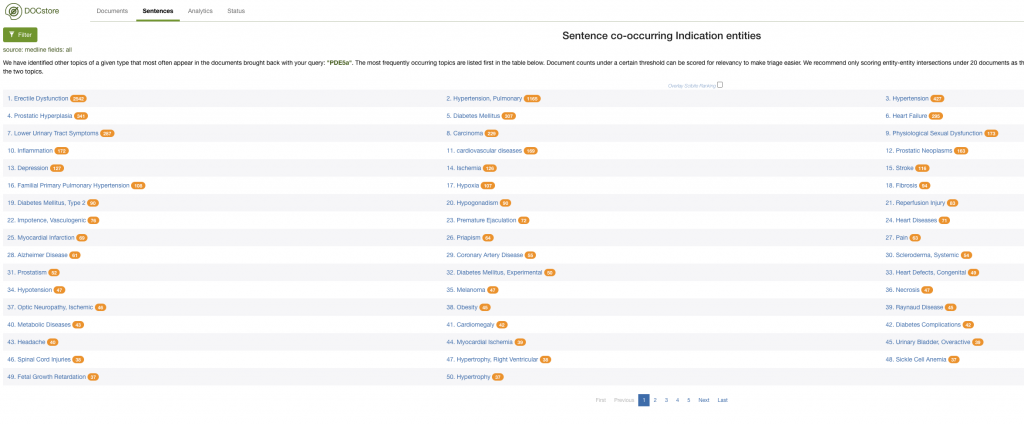
Additionally, you could examine the co-occurrence data to get a feel for the landscape. In this example, I could look at the indications commonly associated with documents mentioning PDE5A:
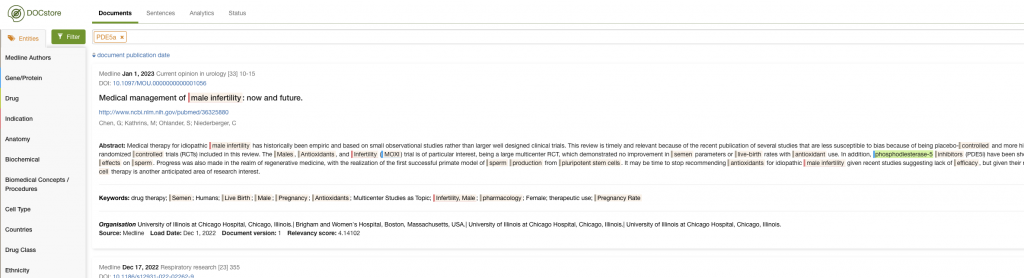 Here, we quickly see that Erectile Dysfunction and Pulmonary Hypotension are associated with PDE5A – and also how much time this can save when working in drug repurposing.
Here, we quickly see that Erectile Dysfunction and Pulmonary Hypotension are associated with PDE5A – and also how much time this can save when working in drug repurposing.
You could also look at co-occurrences on a sentence level. Sentence level co-occurrences are stronger indicators of a real association between entities than document level. Why? Because at a document level you might find entities in keywords section that hold spurious and unrelated terms.
 A comprehensive autocomplete index helps guide your searches. A little bit more in depth than GSK the company or GSK the protein!
A comprehensive autocomplete index helps guide your searches. A little bit more in depth than GSK the company or GSK the protein!
Fit-For-Science Search
But you’re not limited to entities and types that have already been curated. You can build your own vocabularies or use plain text.
Remember that semantically enabled search is as good as the vocabularies it’s built on. An excellent vocabulary with a huge number of synonyms means that typing in the brand name of a drug also brings up papers associated with its clinical name.
And there you have it – pitted against the depth and breadth that semantic search offers, keyword search simply cannot compete in terms of accuracy, full awareness, or efficiency. Semantic search allows you to buy back valuable time that would otherwise be spent sifting through huge amounts of documents, and even convert textual data into something you can integrate across your systems, thanks to entity recognition.
Ready to learn more? Check out:
- Benefits of Semantic Enrichment Across the Drug Development Pipeline
- The Evolution and Importance of Biomedical Ontologies for Scientific Literature.
RightFind Insight, powered by the SciBite® platform, brings the power of semantic enrichment to the search and reading experience to turn information into knowledge and accelerate new discoveries. Learn more here.


